| Title |
Characterization of the retroviral budding factor ALIX |
| Publication Type |
dissertation |
| School or College |
School of Medicine |
| Department |
Biochemistry |
| Author |
Landesman, Michael Benjamin |
| Date |
2012-05 |
| Description |
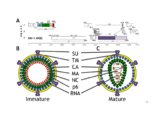
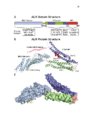
Retroviral virion assembly and budding is orchestrated by the structuralGag polyprotein. To facilitate budding, Gag proteins encode peptide motifs, termed late assembly domains that recruit components of the cellularESCRT pathway, including TSG101 and ALIX. In cells, the ESCRT pathwaymediates membrane fission events required for intralumenal endosomal vesicleformation and for abscission during cytokinesis. Many enveloped viruses, haveevolved to usurp this endogenous membrane fission activity to bud from hostcells. The goal of my dissertation research was to understand how the ESCRTpathway functions in retrovirus budding, with a particular emphasis oncharacterizing the ESCRT-‐associated ALIX protein. ALIX comprises an N-‐terminal Bro1 domain, a central V domain, and a C-‐ terminal proline-‐rich region (PRR). My research has elucidated new roles for each of these regions. Chapter 4 expands our understanding of Bro1 domain structure and function. The crystal structure of the Bro1-‐domain containing protein BROX is presented and compared to structures of Bro1 domains from human ALIX and yeast Bro1p. Using viral budding assays, the importance of two conserved hydrophobic patches was confirmed, and I established the functional requirement for a unique and highly exposed phenylalanine residue (Phe105) onthe convex surface of the ALIX Bro1 domain. Gag proteins from the human immunodeficiency virus type-‐1 (HIV-‐1) and from the equine infectious anemia virus (EIAV) bind to the ALIX V domain through a tyrosine-‐based peptide late assembly domain sequences whose consensus amino acid sequence is YPXL. However, some strains of the simian immunodeficiency virus (SIV) recruit ALIX, yet lack a consensus YPXL motif. My work with SIV in Chapter 2, reveals an expanded family of late domains that bind to ALIX to facilitate virion release. Chapter 3 describes research supporting a multistate model for ALIX protein activation. We show that the PRR of ALIX folds back against the upstream domains and auto-‐inhibits V domain binding to viral late domains.Mutations that destabilize a closed V domain conformation repartitioned ALIXinto membrane-‐containing fractions and enhanced virus budding. Theseobservations support a model in which ALIX activation requires dissociation ofthe autoinhibitory PRR, opening of the V domain, and protein dimerization. |
| Type |
Text |
| Publisher |
University of Utah |
| Subject |
pure sciences; biological sciences; activation; budding; HIV; protein activation |
| Subject MESH |
Gene Products, gag; Polyproteins; Virion; Cytokinesis; Virus Assembly; Virus Release; Endosomal Sorting Complexes Required for Transport; Virus Release; Retroviridae; Human Immunodeficiency Virus Proteins; HIV-1; Acquired Immunodeficiency Syndrome; Endosomes; Protein Conformation; Cell Membrane; Saccharomyces cerevisiae Proteins; Yeast, Dried |
| Dissertation Institution |
University of Utah |
| Dissertation Name |
Doctor of Philosophy |
| Language |
eng |
| Relation is Version of |
Digital reproduction of Characterization of the Retroviral Budding Factor ALIX. Print version available at J.Willard Marriott Library Special Collections. |
| Rights Management |
© Michael Benjamin Landesman |
| Format |
application/pdf |
| Format Medium |
application/pdf |
| Format Extent |
8,051,014 bytes |
| ARK |
ark:/87278/s6k39g1g |
| Setname |
ir_etd |
| ID |
1199274 |
| Reference URL |
https://collections.lib.utah.edu/ark:/87278/s6k39g1g |