| Description |
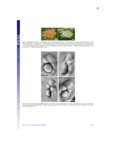
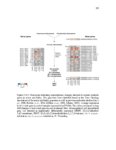
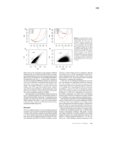
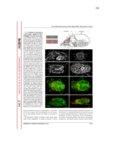
Plant-herbivore interactions have shaped organismal evolution for hundreds of millions of years. For plants, selection has led to the development of physical structures and chemical compounds aimed at deterring animals from feeding, while for herbivores detoxification genes, physical adaptations and behavioral changes have all arisen to circumvent these defenses. The dynamic nature of these interactions is responsible for much of the vast radiation seen in plant and animal species, and nowhere among metazoans is the radiation more profound than among the arthropods. From generalist species capable of feeding on dozens of plant families, to extreme specialists only able to feed on one host, arthropod species span the gamut of herbivore life histories. This work documents advances made in the study of two herbivorous arthropods at opposite ends of the generalist to specialist spectrum: Tetranychcus urticae, the cosmopolitan spider mite capable of feeding on over 1,100 plant hosts, and Aculops lycopersici, the tiny specialized tomato russet mite. For T. urticae, the expansion of chemoreceptor genes to levels higher than any other arthropod species is documented, along with research analyzing the genes responsible for synthesizing carotenoids, which play a key role in enabling this mite to overwinter in temperate climates. For A. lycopersici, the compact genome of this mite is presented, providing the first example of a sequenced Eriophyid genome and shedding light not only on the genomic architecture of long-term host specialization, but also the mechanisms driving extreme genome reduction. From the plant side, this work documents the transcriptomic response of two commercially important crops, barley and maize, to feeding by the generalist T. urticae and the more specialized Banks grass mite, as well as to artificially induced wounding over a 24-hour time course; this allows the host responses to generalist and specialist herbivores, and how this differs from the physical damage they cause, to be investigated. Finally, work to uncover the basis of expression quantitative trait loci underlying ribosomal RNA expression in the model organism Arabidopsis thaliana is documented, along with genetic map improvements made through the identification of structural variants and their link to gene expression and certain phenotypes. |